Introduction and Workflow
- ATAC-seq identifies open chromatin regions, marking transcriptionally active areas in the genome, and provides insights into chromatin accessibility and gene expression regulation.
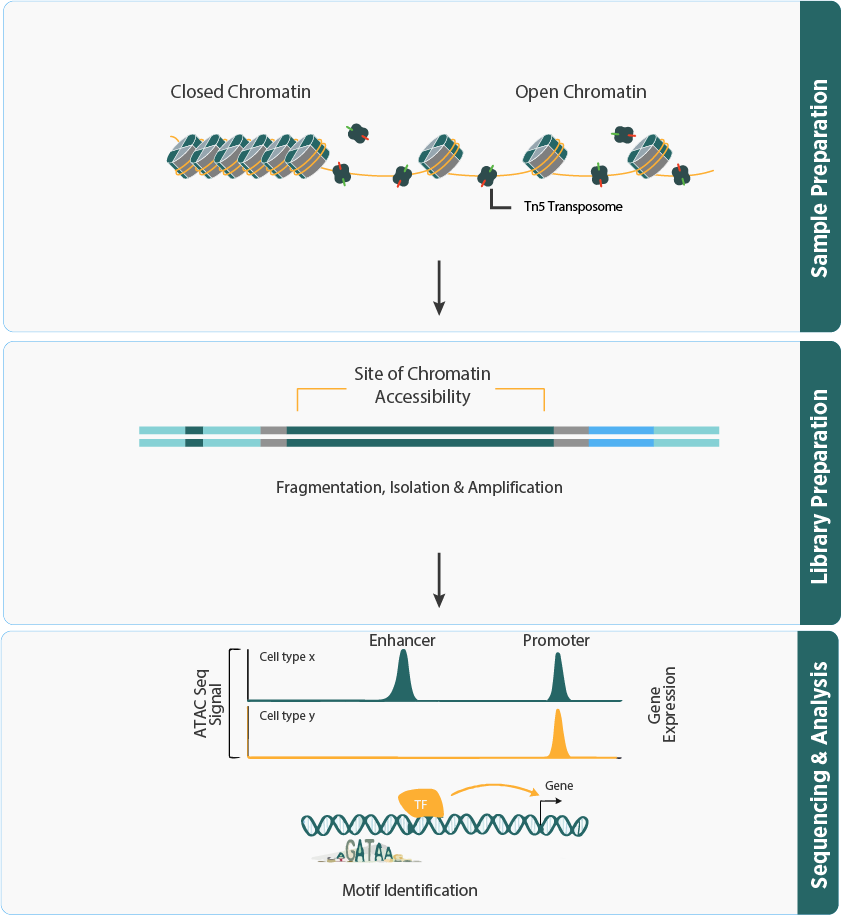
- The workflow uses Tn5 transposase to tag and amplify open chromatin (transposase-accessible) regions, followed by DNA fragmentation, purification, library construction, and sequencing.
- The sequenced fragments are analysed using advanced bioinformatics tools to characterize the regulatory landscape, including chromatin accessibility, nucleosome mapping, and transcription factor binding.
- Assist in numerous applications like biomarker discovery, identification of novel enhancer, analysis of cell-type specific regulation, evolutionary studies and comparative epigenomic studies.
Advantage
- ATAC-seq has become the method of choice for studying chromatin accessibility due to its efficiency and robust performance.
- Requires only 50,000 cells per sample, offering high sensitivity and making it ideal for studies with limited cell populations. This technique is versatile, applicable to both bulk tissue and single-cell analysis.
- Features simplified experimental steps, offering good reproducibility and a high success rate in generating reliable data.
- Simultaneously reveals the genomic locations of open chromatin, DNA-binding proteins, and transcription factor binding site interactions, providing a holistic view of chromatin accessibility.
Applications of ATAC Sequencing
- Chromatin Accessibility and Epigenetic Mapping- ATAC sequencing enables the identification of open chromatin regions, offering insights into regulatory elements like promoters, enhancers, and transcription factor binding sites. This approach is essential for understanding gene regulation and studying the epigenetic landscape across different cell types, tissues, and conditions.
- Cancer Genomics and Neuroscience- ATAC sequencing plays a crucial role in cancer research by identifying alterations in chromatin accessibility that drive tumor development or progression. It helps uncover cancer-specific regulatory elements and potential therapeutic targets. Additionally, in neuroscience, ATAC sequencing is used to investigate the chromatin landscape of neural cells and tissues.
- Cell Type Identification and Heterogeneity- When applied at the single-cell level, ATAC sequencing can reveal chromatin accessibility patterns in individual cells. This capability allows for the identification of distinct cell types, the study of cellular heterogeneity, and an understanding of how chromatin accessibility varies among different cell populations within a tissue.
- Transcription Factor Binding and Developmental Biology- ATAC sequencing, combined with transcription factor motif analysis, helps predict and validate transcription factor binding sites, elucidating their roles in regulating gene expression. This is particularly valuable in developmental biology, where ATAC sequencing is employed to study chromatin dynamics over time.
Service Specifications
Sample Requirement
Cell sample ≥ 50,000 cells; Nucleus sample ≥ 50,000 nuclei
Please refer to sample submission guidelines or Contact Us!
Sequencing Platform
Illumina NovaSeq 6000/ NovaSeq X

Deliverables
- The original sequencing data
- Experimental results
- Bioinformatics and Data Analysis Report
- Details of ATAC Sequencing (customizable)
