Degradome Sequencing
Introduction and Workflow
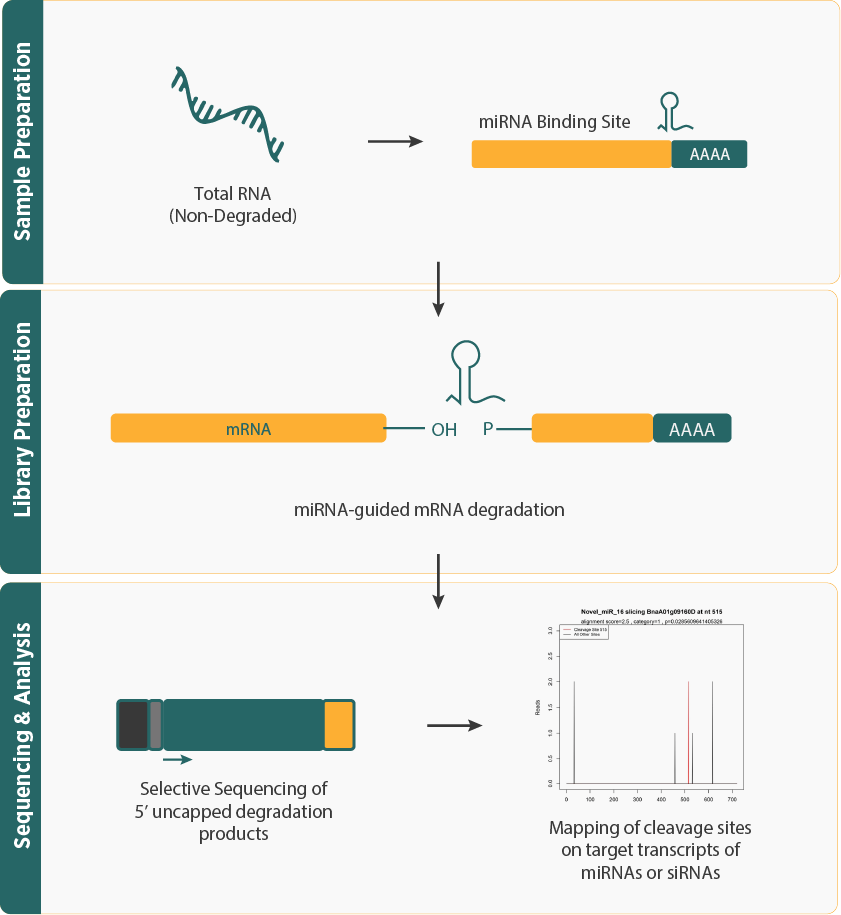
- Degradome sequencing, also called Parallel Analysis of RNA Ends (PARE), is a high-throughput technique to analyze RNA degradation products, uncovering RNA stability, miRNA targets, and regulatory networks.
- Degradome Sequencing Workflow combines next-generation sequencing (NGS) with a modified 5' RACE method, involving RNA extraction, adapter ligation to capture degradation products, cDNA synthesis, and high-throughput sequencing to map RNA cleavage sites.
- Bioinformatics analysis for Degradome Sequencing include data preprocessing, aligning reads to reference genomes, identifying miRNA/ta-siRNA cleavage sites, and quantifying cleaved targets, revealing RNA decay dynamics and regulatory mechanisms.
- The method facilitates the discovery of novel miRNAs and ta-siRNAs, studies on miRNA-mediated gene regulation, and comparative RNA decay analysis across species, contributing to a comprehensive understanding of RNA life cycles.
Advantage
- Degradome sequencing provides a focused analysis of RNA degradation, specifically targeting cleavage sites mediated by miRNAs and ta-siRNAs, making it ideal for studying gene regulation and RNA stability.
- By targeting degradation products, degradome sequencing minimizes the impact of intact RNAs, enhancing sensitivity for detecting miRNA targets and cleavage events.
- Degradome sequencing focuses on RNA cleavage sites, eliminating the need for additional steps like ribosomal RNA removal, making it a more cost-effective alternative to broader transcriptome studies.
- Degradome sequencing enables the identification of novel regulatory elements, such as circRNAs and biomarkers, expanding its utility in understanding RNA regulatory networks.
Bioinformatics Pipeline

Applications of Degradome Sequencing
- Gene Regulation Studies- Used to identify miRNA targets and cleavage sites, elucidating the regulatory networks that control gene expression and RNA turnover in different organisms.
- Developmental Biology- Explores RNA degradation patterns during developmental stages, uncovering the molecular mechanisms underlying cell differentiation and tissue formation.
- Disease Research- Investigates RNA cleavage profiles associated with diseases such as cancer and neurodegenerative disorders, helping to identify potential biomarkers and therapeutic targets.
- Functional Genomics- Provides a detailed view of RNA decay dynamics, enabling the study of gene function and the roles of regulatory RNAs in cellular processes.
- Comparative Genomics- Analyzes RNA degradation patterns across species or environmental conditions to understand evolutionary conservation and differences in regulatory mechanisms.
- Stress and Environmental Responses- Examines changes in RNA stability under stress conditions, shedding light on the molecular mechanisms of stress adaptation and survival strategies.
Service Specifications
Sample Requirement
Sample type: Total RNA without degradation or DNA contamination
Total RNA≥ 20 μg, Minimum Quantity: 15 μg, Concentration≥ 100 ng/µL
Please refer to sample submission guidelines or Contact Us!
Sequencing Platform
Illumina NovaSeq 6000/ NovaSeq X

Deliverables
- The original sequencing data
- Experimental results
- Bioinformatics and Data analysis report
- Details of Degradome Sequencing (customizable)
