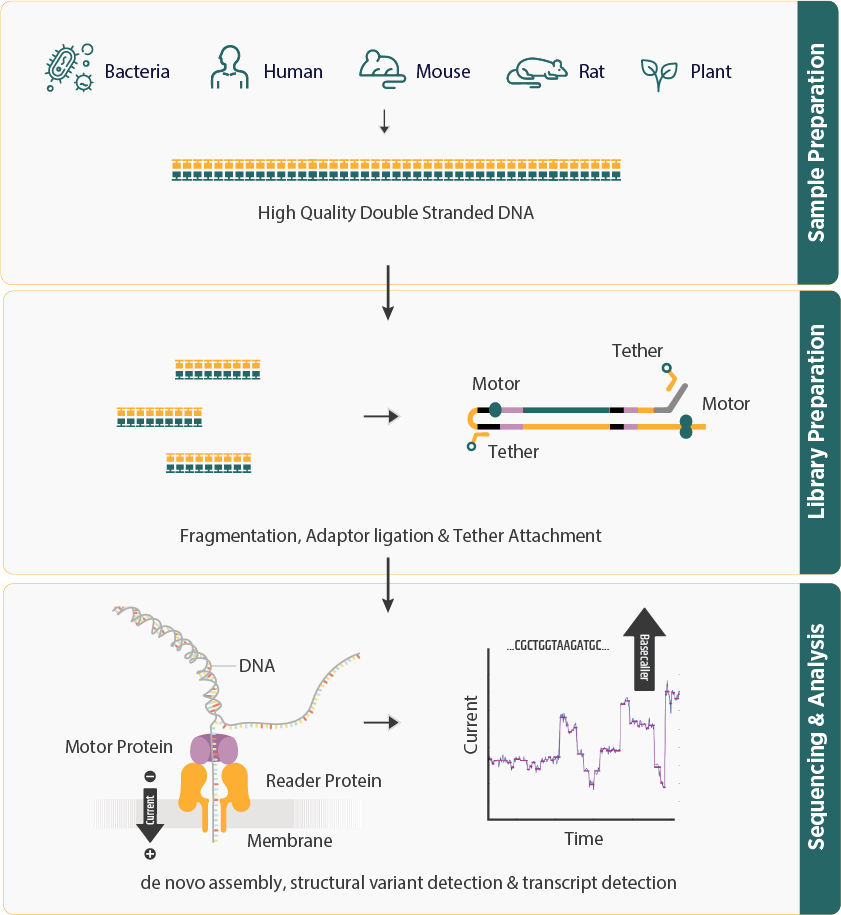
Introduction and Workflow
- Whole Genome Long Read Sequencing captures an organism's entire genome in a single read using extended strands, allowing precise analysis of complex genomic regions and structural variations.
- Technologies like Oxford Nanopore and PacBio enable real-time sequencing with high accuracy. The process involves DNA isolation, fragmentation, and advanced bioinformatics analysis for accurate variant detection.
- This workflow supports sequencing for humans, animals, plants, bacteria, and disease-related microbes using long continuous reads for better genome assembly.
Advantage
- Captures long DNA strands in a single read, providing complete and accurate genome representation.
- Reduces gaps and errors, enabling precise detection of large structural variants.
- Facilitates high-quality de novo genome assembly.
- Real-time sequencing allows faster insights and decision-making, especially in clinical settings.
Comparison of Sequencers
Comparison and specification of sequencing platforms between short-read and long-read sequencing on WGS
Different sequencing platforms have unique characteristics and applications. The comparison below highlights key differences:
| Platform Types | PacBio Sequel II/IIe | Nanopore PromethION |
|---|---|---|
| Read Length | Average ≥ 15 kb
bp to kb | Average > 17 kb
Kb to Mb |
| Variant Calling | Long Read Length with High Accuracy: Good coverage of highly repetition and complexity area
Detect SVs with high precision | Long Read Length with High Accuracy: Good coverage of highly repetition and complexity area
Detect SVs with high precision |
| Amplification/GC Bias | NO GC Bias & Amplification Bias
PCR-free to allow a uniform coverage and High Contiguity | NO GC Bias & Amplification Bias
PCR-free to allow a uniform coverage and High Contiguity |
| Machine Errors | Relaxed requirement for cycle efficiency | Relaxed requirement for cycle efficiency |
| DNA Methylation Detection | Simultaneous Epigenetic Characterization
Diverse DNA methylation types: CpG, CHH, CHG, 6mA, 4mC, 5hmc | Simultaneous Epigenetic Characterization
Diverse DNA methylation types: CpG, CHH, CHG, 6mA, 4mC, 5hmc |
Long Read Sequencing Technologies
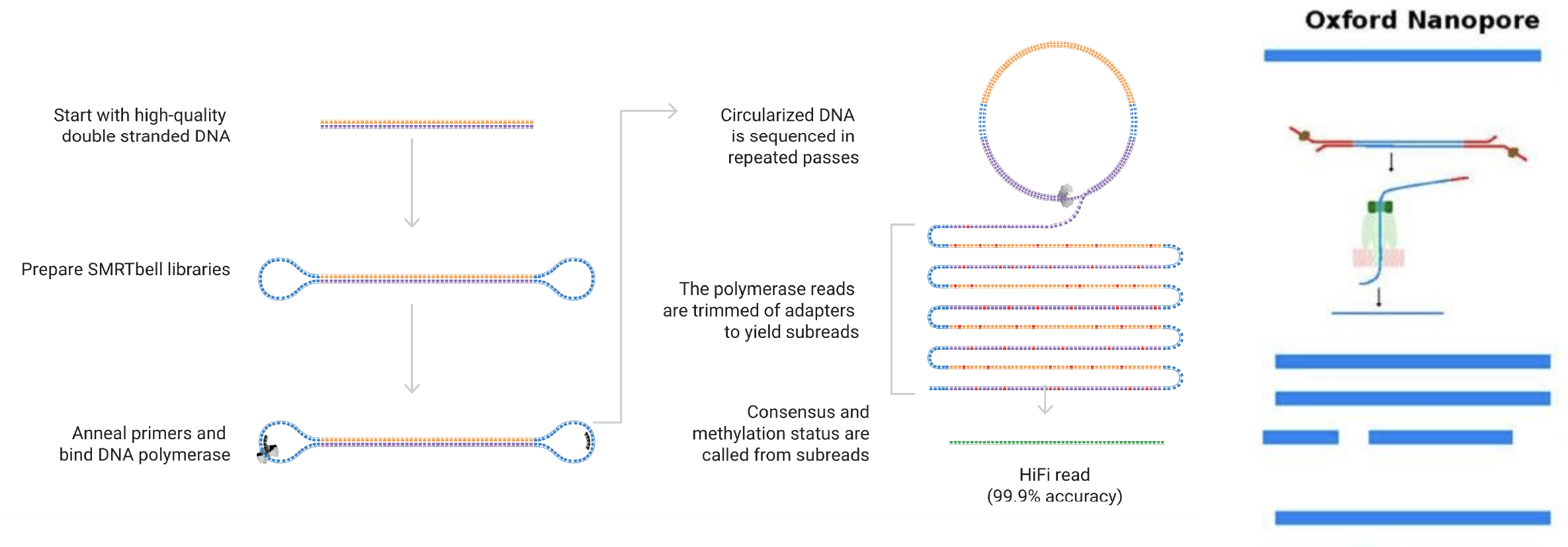
Long Read Sequencing using Oxford Nanopore Technology
OXFORD NANOPORE
Real-time DNA Analysis
- Long-read Sequencing using Oxford Nanopore Technologies (ONT) delivers longer, continuous, and unambiguously assembled sequences, resulting in fewer contigs and enhancing overlap for accurate genome assembly.
- Helps in sequencing long stretches of DNA for complete genome assemblies of microbial, human, animal and plant species.
- Enables direct, real-time analysis of long DNA or RNA fragments by measuring changes in electric current as they pass through a nanopore embedded in a flow cell.
- Nanopore sequencing offers advantages across multiple research areas, including genome assembly, full-length transcript detection, base modification detection, and specialized applications like rapid clinical diagnoses and outbreak surveillance.
Long Read Sequencing using PacBio Technology
- PCR-free SMRT technology producing Hi-Fi reads via circular consensus sequencing (CCS) mode, yielding long reads up to 25 kb with 99.9% base level accuracy.
- Enables rapid and cost-effective generation of contiguous, complete, and accurate de novo genome assemblies, even for complex genomes.
- Allows the haplotype resolution of complex polyploids, particularly in plants.
- The technology can be utilized to provide a comprehensive view of the epigenome and transcriptome, as well as uncover different variants such as SNPs, homopolymer repeats, translocations, structural variants, and copy number variants.
PACBIO SMRT
Hi-Fi Circular Consensus Sequencing
Applications of Long Read Sequencing
- Structural Variant Detection- Identifies large insertions, deletions, duplications, inversions, and translocations that short reads often miss.
- De Novo Genome Assembly- Generates complete genome assemblies even in repetitive or uncharted genomic regions.
- Haplotype Phasing- Spans long stretches of DNA, useful for understanding inherited traits and allelic relationships.
- Complex Region Analysis- Effective in resolving telomeres, centromeres, MHC, and other challenging genomic regions.
Service Specifications
Sample Requirement
Genomic DNA, Cultivated cells, Blood, tissues, and other samples
Please refer to sample submission guidelines or Contact Us!
Sequencing Platform
PacBio, Oxford Nanopore

Deliverables
- The original sequencing data
- Experimental results
- Bioinformatics and Data Analysis Report
- Details of Whole Genome ReSequencing (customizable)
