Introduction and Workflow
- Whole Genome Denovo Sequencing involves sequencing an organism's entire genome from scratch, without a reference genome. This approach is essential for species with unsequenced or incomplete genomes.
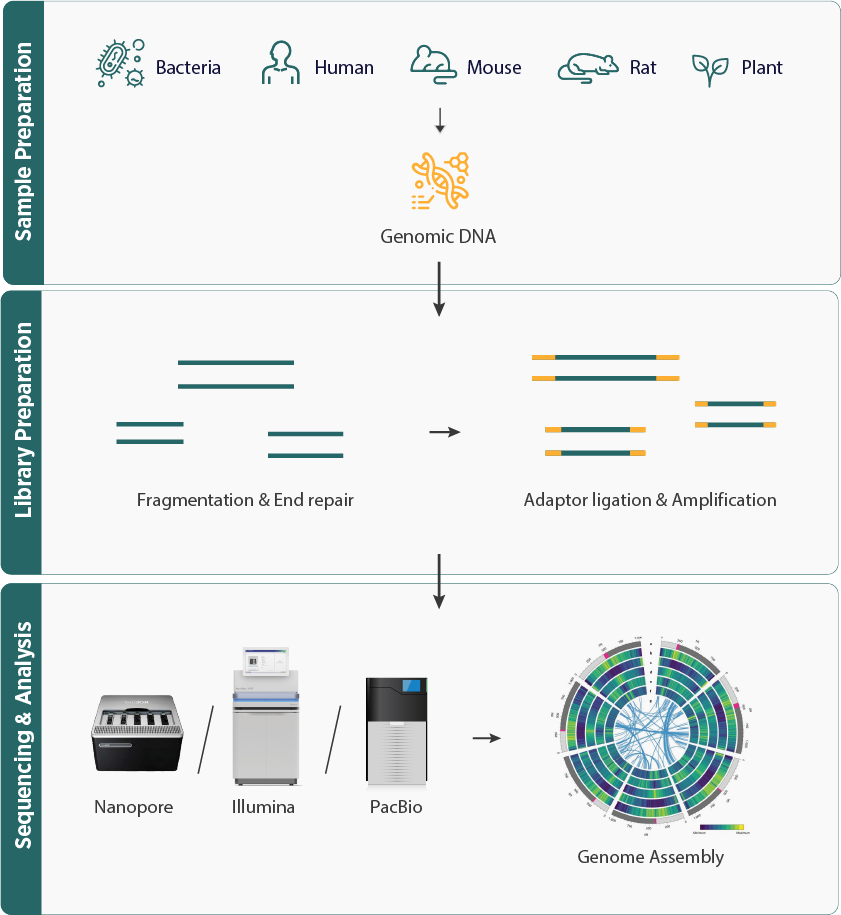
- The workflow includes isolating DNA, fragmenting it, and sequencing to produce millions of short reads.
- These reads are then assembled into longer sequences, called contigs, using bioinformatics tools in the genome assembly process.
- It can be used for sequencing diverse species, such as agriculturally important livestock, plants, bacteria, or disease-related microbes.
Advantage
- Eliminates the need for pre-existing reference genomes, which allows for the discovery of novel genetic elements and variations that may not be present in other genomes, providing a truly unbiased view of the genome.
- Identifies new genes and genetic variations that may be missed by relying on reference genomes alone.
- Offers high-resolution insights into the genome, including complex regions such as repetitive sequences and heterochromatic regions, which are often challenging to assemble with other sequencing approaches.
- Enhances our understanding of genetic diversity, facilitates comparative genomics, and contributes significantly to advancements in genomic research across various fields.
Bioinformatics Pipeline

Applications of Whole Genome Denovo Sequencing
- Genome Assembly- Enables the construction of comprehensive and accurate genome assemblies from scratch, essential for understanding the genetic blueprint of organisms where reference genomes are unavailable or incomplete.
- Genetic Variation Analysis- Facilitates the identification of genetic variants, including single nucleotide polymorphisms (SNPs), insertions, deletions, and structural variants, which are crucial for studying genetic diversity, evolution, and disease.
- Functional Genomics- Supports the annotation of genes, regulatory elements, and non-coding regions, providing insights into gene function, gene expression regulation, and the roles of various genomic elements in biological processes.
- Comparative Genomics- Allows for the comparison of genome sequences across different species or strains, aiding in the identification of evolutionary relationships, adaptation mechanisms, and the conservation of key genomic features.
- Applications in Diverse Fields- Supports various research applications such as identifying genetic variants associated with diseases, exploring evolutionary relationships, and understanding genetic diversity in populations, making it valuable for personalized medicine, evolutionary studies, and crop improvement.
Service Specifications
Sample Requirement
Genomic DNA, Cultivated cells, Blood, tissues, and other samples.
Please refer to sample submission guidelines or Contact Us!
Sequencing Platform
Illumina NovaSeq 6000/ NovaSeq X

Deliverables
- The original sequencing data
- Experimental results
- Bioinformatics and Data Analysis Report
- Details of Whole Genome Denovo Sequencing (customizable)
