LncRNA Sequencing
Introduction and Workflow
- Long non-coding RNAs (lncRNAs) are a class of RNA molecules longer than 200 nucleotides that do not code for proteins. Based on their position and direction of transcription, lncRNAs are further classified into subtypes such as antisense, intergenic, overlapping, intronic, bidirectional, and processed. These RNAs play essential roles in regulating gene expression at various levels, impacting a wide range of cellular processes.
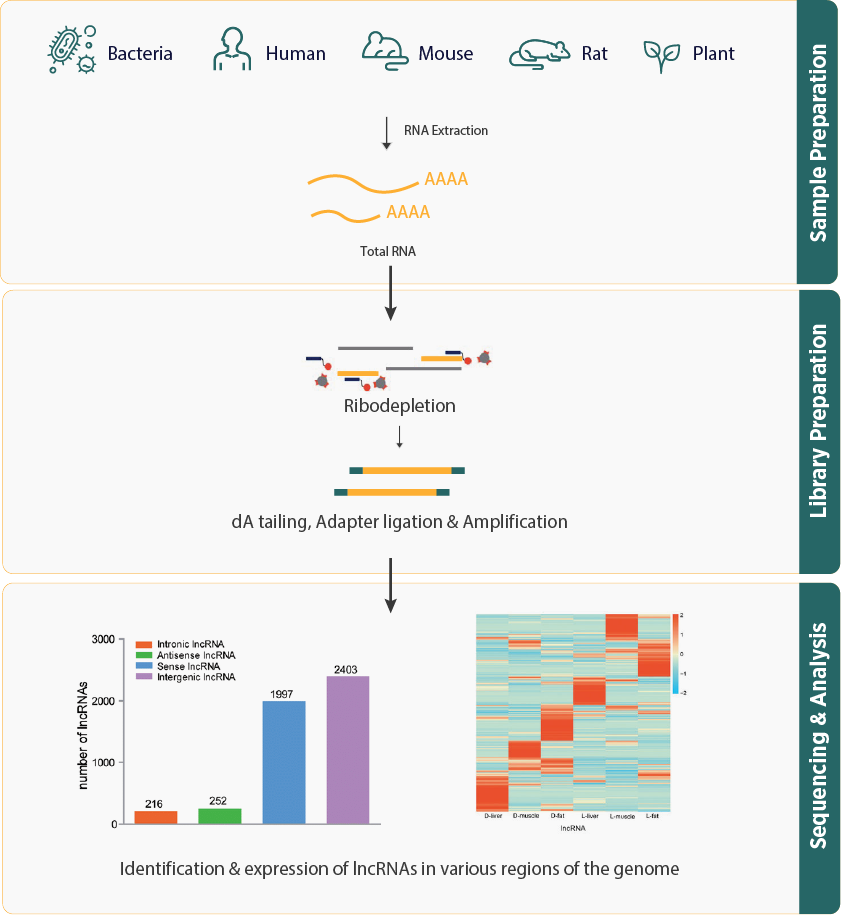
- lncRNA sequencing workflow starts with the removal of ribosomal RNA (rRNA) from the total RNA sample to isolate lncRNAs and mRNAs. The isolated RNA is then subjected to library preparation and high-throughput sequencing. Through specific computational pipelines lncRNAs will be identified during bio-informatic analysis.
- The bioinformatics pipeline for lncRNA sequencing involves the alignment of sequencing data to reference genomes, identification of lncRNAs, quantification of their expression level, and other downstream analyses.
- lncRNA sequencing uncovers their roles in gene regulation and highlights their various functions, including chromatin remodeling, transcriptional control, nucleosome repositioning, and post-transcriptional processing.
Advantage
- Identifies both well-known and novel lncRNAs, providing fresh insights into their roles and potential as biomarkers.
- Reveals lncRNAs' involvement in a broad range of molecular functions, such as chromatin remodeling, which is essential for regulating gene expression. This process includes modifying chromatin structure to control gene accessibility and influencing various cellular mechanisms, demonstrating lncRNAs' crucial roles in gene regulation and cellular function across mammals and plants.
- Provides a thorough understanding of lncRNAs' roles in gene regulation, encompassing both transcriptional and post-transcriptional levels. It highlights how lncRNAs affect transcription factors, alter nucleosome positioning, and contribute to RNA splicing and stability, thus impacting overall gene expression and cellular processes.
- Enables high-resolution analysis, allowing for precise detection and detailed characterization of lncRNA expression and their regulatory networks, offering a deep dive into their functional roles within cells.
Bioinformatics Pipeline

Applications of lncRNA Sequencing
- LncRNA sequencing is widely utilized across plants, animals, microbes, and humans.
- Cancer Research- Identifies tumor-specific lncRNAs and their roles in cancer development, progression, and metastasis, offering potential biomarkers and therapeutic targets.
- Developmental Biology- Studies lncRNAs involved in developmental processes and cell differentiation, providing insights into gene regulation during development.
- Disease Mechanisms- Uncovers lncRNAs associated with various diseases, including genetic disorders, cardiovascular diseases, and neurodegenerative conditions, facilitating the discovery of novel diagnostic and prognostic markers.
- Drug Discovery- Assists in identifying lncRNAs that can be targeted or modulated by drugs, contributing to the development of new therapeutic strategies.
- Gene Regulation- Explores lncRNAs' roles in regulating gene expression through chromatin remodeling, transcriptional control, and post-transcriptional modifications, enhancing the understanding of complex gene regulatory networks.
- Functional Genomics- Provides insights into the functional roles of lncRNAs in cellular processes, helping to elucidate their impact on gene expression and cellular functions.
- Comparative Genomics- Analyzes lncRNA profiles across different species or conditions, supporting research into evolutionary differences and functional conservation of lncRNAs.
Service Specifications
Sample Requirement
Total RNA ≥ 1 μg, Cells≥ 2×106, or Tissue ≥ 500 mg.
Please refer to sample submission guidelines or Contact Us!
Sequencing Platform
Illumina NovaSeq 6000/ NovaSeq X

Deliverables
- The original sequencing data
- Experimental results
- Bioinformatics and Data analysis report
- Details of LncRNA Sequencing (customizable)
