Small RNA (sRNA) sequencing
Introduction and Workflow
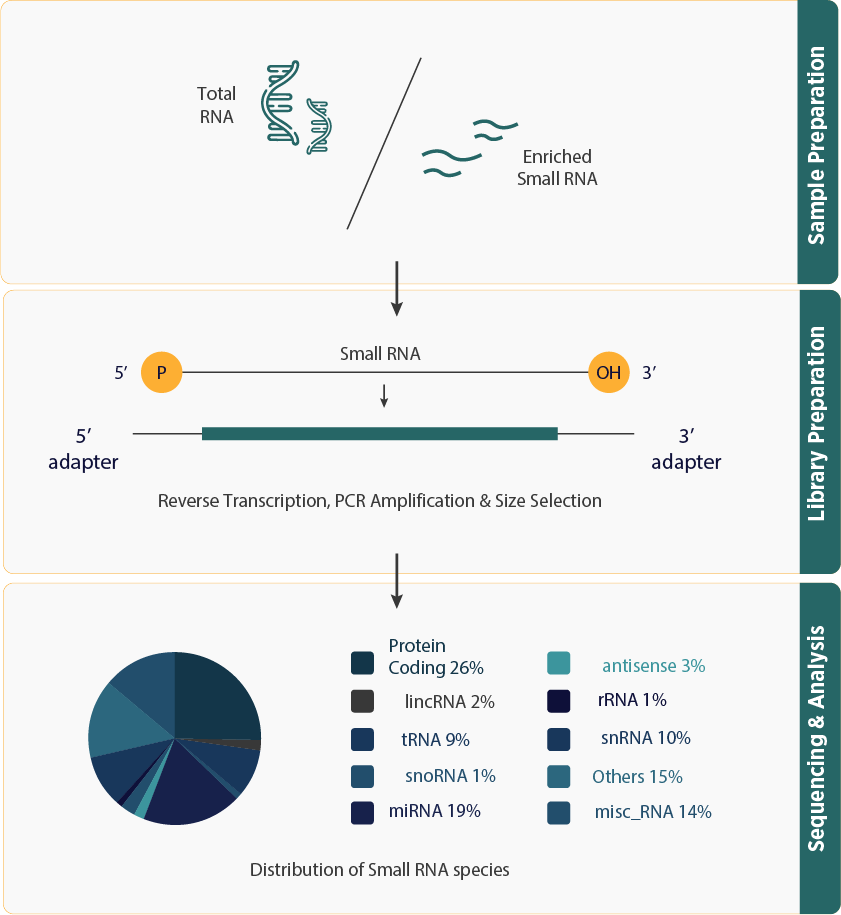
- Small RNA sequencing (sRNA-seq) is designed to identify and analyze small non-coding RNAs (ncRNAs) that are under 200 nucleotides in length. These small RNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs), small nucleolar RNAs (snoRNAs), PIWI-interacting RNAs (piRNAs), and trans-acting siRNAs (tasiRNAs), play crucial roles in gene silencing and post-transcriptional regulation.
- The sRNA-seq workflow involves isolating miRNAs from samples, followed by preparing libraries with specific adapters and sequencing them using high-throughput platforms.
- The bioinformatics pipeline for sRNA-seq encompasses data preprocessing, alignment with reference genomes, and identification of small RNA species. This analysis includes differential expression studies, characterization of novel small RNAs, and pathway analysis to gain functional insights into sRNA-target interactions.
- Facilitates the discovery of novel small RNAs and offers detailed insights into their expression and functions. It links post-transcriptional regulation to phenotypic outcomes, aiding in the understanding of disease mechanisms and the identification of new biomarkers.
Advantage
- Provides a comprehensive view of the small RNA landscape by capturing a broad range of small RNA species, including miRNAs, siRNAs, snoRNAs, piRNAs, and tasiRNAs.
- Facilitates comparative studies by allowing the comparison of small RNA expression profiles across different conditions or species, supporting investigations into developmental, environmental, and disease-related changes.
- Identifies previously unmapped small RNAs and isoforms, as well as novel biomarkers, expanding the scope of genetic research and potential clinical applications.
- Enhances understanding of how post-transcriptional regulation affects phenotypes and offers detailed functional insights into small RNA-target interactions and their roles in various cellular pathways, improving the understanding of gene regulation and function.
Bioinformatics Pipeline

Applications of Small RNA (sRNA) sequencing
- sRNA sequencing is widely utilized across plants, animals, microbes, and humans.
- Cancer Research- Identifies novel small RNAs and biomarkers, revealing their roles in tumor development and progression, and aiding in the discovery of potential therapeutic targets.
- Developmental Biology- Examines small RNA expression during development to understand gene regulation, cellular differentiation, and developmental disorders.
- Genetic Studies- Analyzes allele-specific expression and variations in small RNAs to gain insights into genetic regulation and variability.
- Microbial Genomics- Investigates small RNA profiles in bacteria, aiding in the understanding of gene regulation, host-pathogen interactions, and antibiotic resistance.
- Neuroscience- Explores small RNA involvement in brain function and neurodevelopmental disorders, offering insights into neural regulation and disease mechanisms.
- Immunology- Studies small RNA expression in immune cells to understand immune responses, autoimmune diseases, and immune system regulation.
- Environmental and Ecological Research- Analyzes the impact of environmental changes on small RNA profiles in plants, animals, and microbes, supporting studies on adaptation and stress responses.
Service Specifications
Sample Requirement
Total RNA ≥ 1 μg, Cells≥ 2×106, or Tissue ≥ 500 mg.
Please refer to sample submission guidelines or Contact Us!
Sequencing Platform
Illumina MiSeq, Nextseq 550

Deliverables
- The original sequencing data
- Experimental results
- Bioinformatics and Data analysis report
- Details in Small RNA Sequencing for your writing (customization)
