Introduction and Workflow
- Method to resolve large-scale structural variations by linearizing strands of high molecular weight (HMW) DNA and imaging them in their native state, offering high sensitivity and accuracy to correct sequencing-based errors.
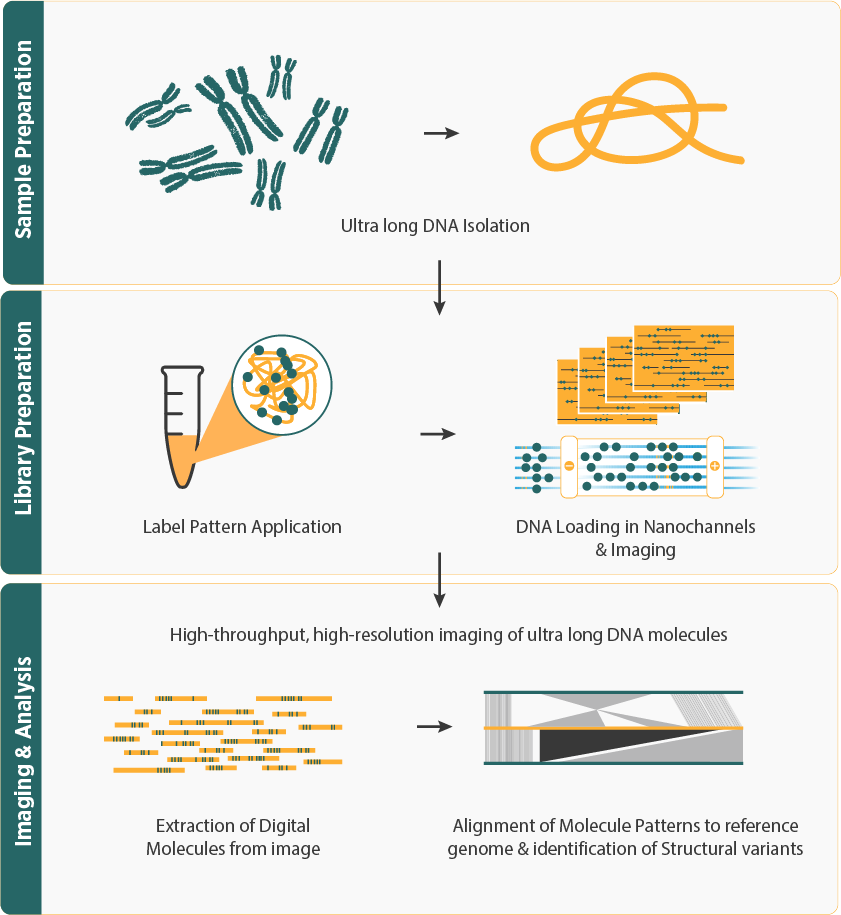
- The workflow begins with isolating high-molecular-weight DNA, followed by labeling specific sequences with fluorescent tags. These labeled long DNA molecules are then loaded onto Saphyr Chip's NanoChannels for imaging, where they are linearized and imaged to create a "barcode" pattern representing the genome's structure.
- The barcode patterns are aligned to a reference genome for assembly with enhanced contiguity using bioinformatics tools to detect structural variations and assemble large genomic segments.
- This allows for the comparison of genomic maps, identification of rearrangements, and integration with other sequencing data for comprehensive genome analysis.
- With the application in clinical and discovery research, the technique is improving genomics assembly, understanding of genetic disease and cancer by detecting CNVs, chromosomal aberrations and structural variants. Enables high-resolution analysis of large eukaryotic genomes and their structural features.
Advantage
- Offers detailed genome-wide mapping of protein-DNA interactions, identifying binding sites of transcription factors, regulators, and other DNA-associated proteins.
- Utilizes specific antibodies to precisely capture and sequence protein-DNA complexes, providing accurate reflections of in vivo binding events.
- Reveals critical insights into gene regulation and chromatin dynamics, aiding in the understanding of gene expression, regulatory mechanisms, and epigenetic modifications.
- Facilitates studies on gene regulation in various contexts, including developmental processes, cancer research, and other biological phenomena, enhancing our understanding of complex biological systems and diseases.
Applications of Optical Mapping
- Transcription Factor Binding Analysis- Identifies and maps the binding sites of transcription factors across the genome, revealing their roles in gene regulation and the modulation of gene expression.
- Histone Modification Mapping- Profiles histone modifications to understand their influence on chromatin structure and gene accessibility, providing insights into epigenetic regulation and chromatin dynamics.
- Regulatory Element Discovery- Uncovers enhancers, silencers, and other regulatory elements that control gene expression, aiding in the identification of key regions involved in gene regulation.
- Epigenetic Research- Studies epigenetic marks associated with various cellular states and processes, such as differentiation and development, to elucidate the mechanisms underlying gene expression changes and disease states.
- Cancer Research- Examines changes in chromatin structure and transcription factor binding in cancer cells, identifying potential biomarkers and therapeutic targets for cancer treatment.
- Developmental Biology- Investigates changes in gene regulation during development, revealing how chromatin modifications and transcription factor binding contribute to cellular differentiation and tissue development.
- Drug Discovery- Provides insights into the effects of drugs on chromatin structure and gene regulation, supporting the identification of new drug targets and the development of therapeutic strategies.
Service Specifications
Sample Requirement
Cell sample ≥ 50,000 cells; Nucleus sample ≥ 50,000 nuclei
Please refer to sample submission guidelines or Contact Us!
Sequencing Platform
Illumina NovaSeq 6000/ NovaSeq X

Deliverables
- The original sequencing data
- Experimental results
- Bioinformatics and Data Analysis Report
- Details of WGBS Sequencing (customizable)
