Single Cell RNA Sequencing
Introduction and Workflow
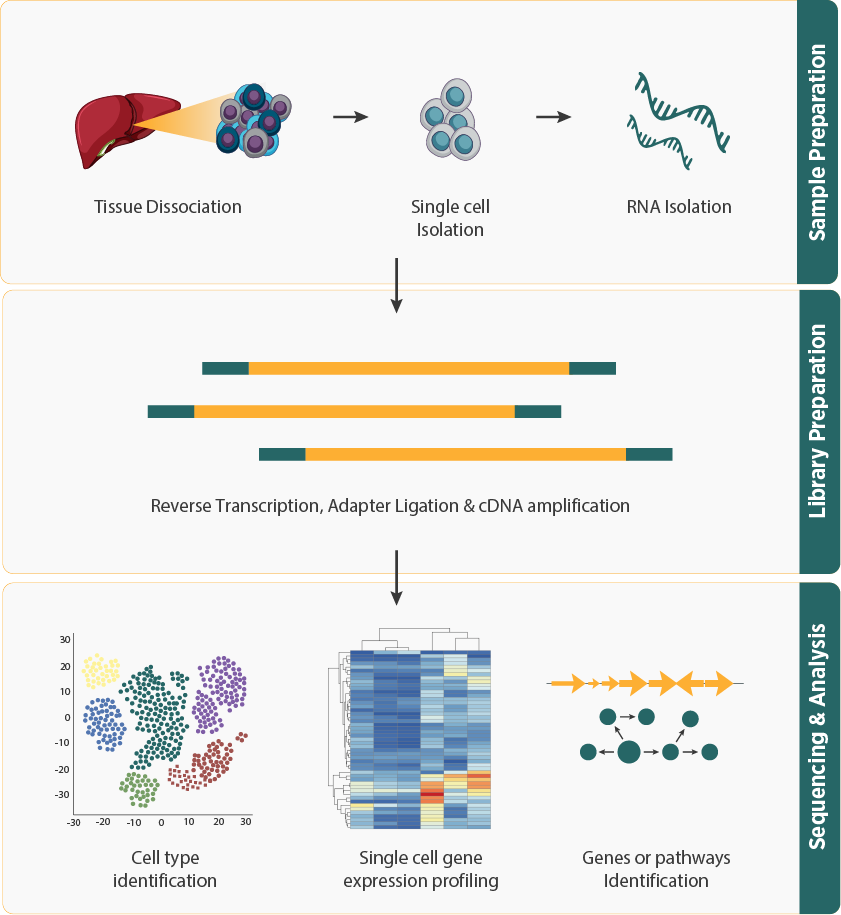
- Single Cell RNA Sequencing (scRNA-seq) analyzes gene expression at the resolution of individual cells, allowing for the exploration of cellular transcriptomic heterogeneity and the discovery of rare cell types within complex tissues.
- The workflow involves isolating individual cells from a sample, often using techniques like fluorescence-activated cell sorting (FACS), followed by RNA extraction, library preparation and (or) all the mRNA molecules in each cell are specifically barcoded either by capturing individual cells in a gel-emulsions (Gem; 10X) or Split-pool approach.
- The bioinformatics pipeline includes data preprocessing, which involves quality control and normalization of the sequencing reads. The reads are then aligned to a reference genome or transcriptome to identify gene expression levels in each cell. Subsequent analysis involves clustering of cells based on expression profiles, differential gene expression analysis, and the identification of cell types and states.
- Single-cell high-resolution transcriptome profiling offers valuable insights into gene regulatory networks and unveils diverse gene expression patterns within individual cells across cell cultures, tissues, and organs.
Advantage
- Unlike traditional bulk RNA sequencing, which averages gene expression across many cells and can hide unique cellular differences, single-cell RNA sequencing examines each cell individually. This reveals variations in gene expression and provides detailed insights into cellular functions, development, and disease mechanisms that are often lost in population-level data.
- Enables the discovery and characterization of rare or previously unrecognized cell populations, essential for understanding complex biological systems and disease mechanisms.
- Delivers high-resolution data on gene expression patterns, allowing for precise mapping of gene regulatory networks and cellular states, and uncovering intricate details of cellular interactions.
- Supports the identification of biomarkers and therapeutic targets by offering a detailed snapshot of gene activity specific to individual cell types or states, facilitating more targeted and effective research and treatments.
Bioinformatics Pipeline

Applications of Single Cell RNA Sequencing
- Single Cell RNA sequencing is widely utilized across plants, animals, and humans.
- Cellular Heterogeneity- Enables the identification and characterization of distinct cell types within complex tissues, providing insights into the diversity and functions of individual cells in health and disease.
- Developmental Biology- Reveals how gene expression changes during development and differentiation, helping to understand the mechanisms driving cell fate decisions and tissue formation.
- Immunology- Provides detailed profiles of immune cell types and states, aiding in the study of immune responses, autoimmune diseases, and vaccine development by uncovering the dynamics of individual immune cells.
- Cancer Research- Helps in identifying rare tumor cell populations, understanding tumor heterogeneity, and discovering novel biomarkers or therapeutic targets specific to different cancer subtypes.
Service Specifications
Sample Requirement
Fresh Viable Samples - Individual Cells for Animal and Protoplasm for Plants
Please refer to sample submission guidelines or Contact Us!
Sequencing Platform
Illumina NovaSeq 6000/ NovaSeq X

Deliverables
- The original sequencing data
- Experimental results
- Bioinformatics and Data Analysis Report
- Details of Single Cell RNA Sequencing (customizable)
