Metatranscriptomics Sequencing
Introduction and Workflow
- Metatranscriptomics sequencing examines RNA transcripts from entire microbial communities within their natural environments during a specific time frame. This approach helps to infer the functional profile of a community under specific conditions, which are usually dependent on the status of the host.
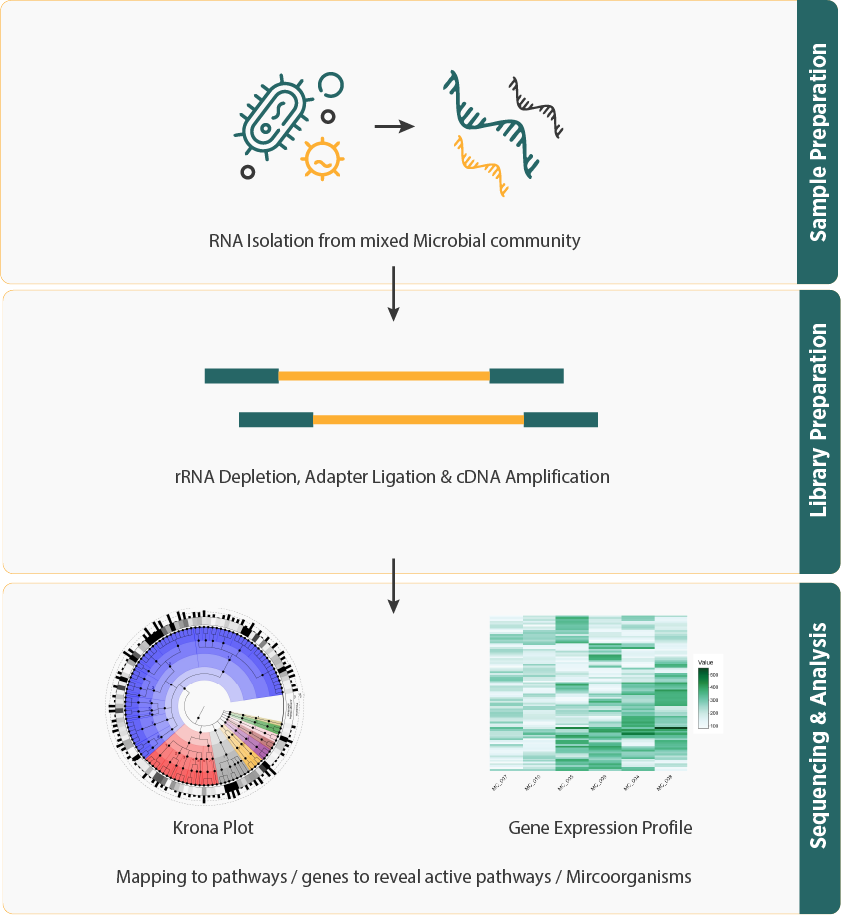
- The process starts with the extraction of total RNA from a mixed microbial sample, without the need for bacterial isolation. Ribosomal RNA (rRNA) is then removed to enrich for mRNA and other non-ribosomal RNAs. The RNA is subsequently processed, prepared into libraries, and sequenced.
- The bioinformatics workflow includes preprocessing the raw sequencing reads, performing quality control, and normalizing the data. The reads are mapped to reference genomes or databases to quantify gene expression levels. Functional annotation and pathway analysis are then carried out to understand the roles of various transcripts and their impact on microbial community functions.
- Metatranscriptomics is applicable to a wide range of environmental samples, such as soil, oceans, and the human gut, facilitating studies on microbial diversity, disease mechanisms, nutrient cycling, and environmental adaptation.
Advantage
- Provides insights into the functional dynamics of microorganisms by revealing active gene expression and their roles within complex communities. This approach uncovers how microorganisms contribute to ecosystem processes and their interactions.
- Analyzes microbial communities directly from environmental samples, removing the need for isolating or cultivating individual microbes. This method captures a more comprehensive view of microbial diversity.
- Uses next-generation sequencing (NGS) to identify novel genes and track transcriptional changes, offering high-resolution data on gene expression. This allows for precise identification of active genes, pathways, and regulatory networks within microbial communities.
- Offers an advantage over metagenomics by including both microbial species information and gene expression data. This dual focus provides a deeper understanding of microbial community changes from a transcriptional perspective and aids in the discovery of potential new genes.
Bioinformatics Pipeline

Applications of Metatranscriptome Sequencing
- Metatranscriptomics sequencing is widely used in microbial genomics to explore microbial diversity, functional potentials, and ecological interactions across environments from soil to human microbiomes.
- Environmental Monitoring- Used to assess microbial community dynamics and functional potential in diverse environments like soil, oceans, and freshwater. It helps track microbial contributions to ecosystem processes and environmental health.
- Biogeochemical Cycling- Investigates the roles of microorganisms in nutrient cycling processes, such as carbon, nitrogen, and sulfur cycles, providing insights into how microbes influence these critical ecological functions.
- Disease Research- Explores the microbial contributions to human health and disease by studying the microbial communities in the human gut, oral cavity, or other body sites, revealing potential biomarkers and therapeutic targets.
- Biotechnology and Industry- Facilitates the discovery of novel enzymes and metabolic pathways from microbial communities, which can be applied in biotechnology, agriculture, and industrial processes.
- Microbial Interactions- Examines interactions between different microorganisms within complex communities, helping to understand symbiotic relationships, competition, and microbial networks.
- Pathogen Detection- Aids in identifying and characterizing pathogenic microbes and their gene expression profiles during infections, enhancing our ability to monitor and manage microbial diseases.
Service Specifications
Sample Requirement
Total RNA samples, isolated from various environmental samples such as soil, water, sediments, roots, leaves, animal gut/intestine etc,.
Please refer to sample submission guidelines or Contact Us!
Sequencing Platform
Illumina NovaSeq 6000/ NovaSeq X

Deliverables
- The original sequencing data
- Experimental results
- Bioinformatics and Data analysis report
- Details of Metatranscriptome Sequencing (customizable)
