Introduction and Workflow
- Hi-C Sequencing is a proximity ligation method that captures the three-dimensional (3-D) structure of chromatin by identifying spatial interactions between different regions of chromosomes.
- It helps understand genome organization, gene regulation, and how chromatin structure affects cellular function.
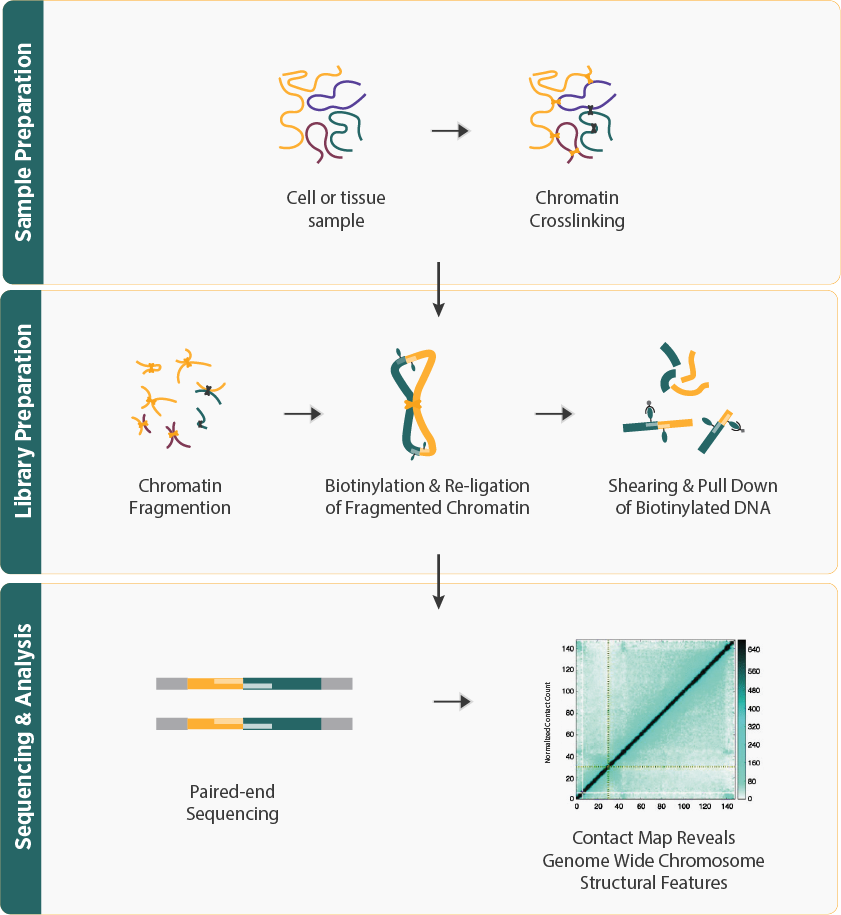
- The workflow involves cross-linking DNA within cells to preserve spatial interactions, followed by fragmentation, ligation of interacting DNA fragments, and sequencing. After sequencing, the resulting data reflects which DNA regions are in close physical proximity within the nucleus.
- The sequenced reads are processed using advanced bioinformatics tools, starting with alignment to a reference genome, followed by the identification of chromatin loops and the construction of 3D models of genome organization.
- Plays pivotal role in multiple applications, including identifying promoter-enhancer interactions for gene regulation studies, detecting structural rearrangements, scaffolding contigs to define chromosomes de novo, and revealing structural errors while accurately reintegrating mis-joined contigs during genome assembly processes.
Advantage
- Quantifies interactions between closely located genomic loci in 3-D space, despite being separated by many nucleotides in the linear genome.
- Provides a detailed view of the 3D architecture of the genome, helping researchers understand how chromatin folding impacts gene regulation, genome organization, and cellular function.
- Captures interactions between distant regions of DNA, enabling the mapping of chromatin loops, topologically associated domains (TADs), and other structural features at high resolution.
- By revealing interactions between regulatory elements, such as enhancers and promoters, Hi-C mapping sheds light on how gene expression is controlled at the epigenetic and transcriptional levels.
Applications of High-throughput Chromosome Conformation Capture (Hi-C) Mapping
- Gene Regulation Studies- Helps in identifying interactions between distant regulatory elements, such as enhancers and promoters, providing insights into gene expression and transcriptional regulation mechanisms.
- Cancer Research- Widely used to study alterations in chromatin structure associated with cancer, aiding in the discovery of genomic rearrangements, fusion genes, and disrupted regulatory networks that contribute to tumorigenesis.
- Developmental Biology- Reveals how genome architecture changes and influences gene expression patterns critical for tissue differentiation and organ development.
- Epigenetics and Disease Mechanisms- Helps in understanding how epigenetic changes, such as histone modifications and DNA methylation, affect chromatin conformation and contribute to diseases like neurodevelopmental disorders, metabolic diseases, and genetic syndromes.
- Comparative Genomics- Allows researchers to compare 3D genome structures across different species, helping to unravel evolutionary changes in genome organization and how they influence biological diversity.
Service Specifications
Sample Requirement
Samples sources including Genomic DNA of human, animals, plants and microorganisms
Please refer to sample submission guidelines or Contact Us!
Sequencing Platform
Illumina NovaSeq 6000/ NovaSeq X

Deliverables
- The original sequencing data
- Experimental results
- Bioinformatics and Data Analysis Report
- Details of Hi-C Sequencing (customizable)
